glmnetlib: A Low-level Library for Regularized Regression
pirls: An R/C++ Package for Penalized, Iteratively Reweighted Least Squares Regression
Goals
Talk: show how many different regression routines (especially glmnet) fit together
Package: write a package that is:
- fast enough to be used
- simple enough to use and understand
- extensible enough to create new routines
Why should we still care about linear models?
Fitting has relatively low complexity
We can figure out how and when they don't work
They are compatible with other learners
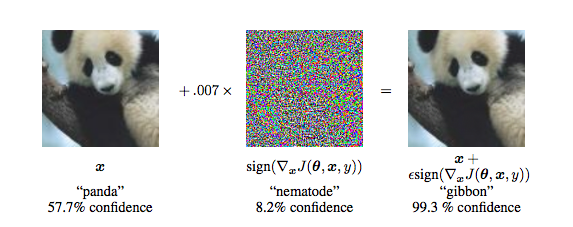
Breaking DL with Adversarial Noise
Figure from Explaining and Harnessing Adversarial Examples by Goodfellow et al.
> train_inds = sample(1:150, 130)
> iris_train = iris[train_inds,]
> iris_test = iris[setdiff(1:150, train_inds),]
> lm_fit = lm(Sepal.Length ~ Sepal.Width+Petal.Length,
+ data=iris_train)
> sd(predict(lm_fit, iris_test) - iris_test$Sepal.Length)
[1] 0.3879703> library(randomForest)
> iris_train$residual = lm_fit$residuals
> iris_train$fitted_values = lm_fit$fitted.values
> rf_fit = randomForest(
+ residual ~ Sepal.Width+Petal.Length+fitted_values,
+ data=iris_train)
>
> lm_pred = predict(lm_fit, iris_test)
> iris_test$fitted_values = lm_pred
> sd(predict(rf_fit, iris_test)+lm_pred -
+ iris_test$Sepal.Length)
[1] 0.3458326> rf_fit2 = randomForest(
+ Sepal.Length ~ Sepal.Width+Petal.Length,
+ data=iris_train)
> sd(predict(rf_fit2, iris_test) - iris_test$Sepal.Length)
[1] 0.40927What is glmnet?
\text{regressor matrix:\ \ } X \in \mathcal{R}^{n \times p}
\text{dependent data:\ \ } y \in \mathcal{R}^n
\text{slope coefficient estimates:\ \ } \hat \beta \in \mathcal{R}^p
\text{penalty parameter:} \lambda \in [0, \infty)
\text{elastic net parameter:} \alpha \in [0, 1]
Notation:
\text{a weight matrix:} W \in R^{n \times n}
\text{a link function:} g
\min_{\hat{\beta}} ||W^{1/2} (y - g(X \hat \beta))||^2 + (1-\alpha) \frac{\lambda}{2} ||\hat \beta||^2 + \alpha \lambda ||\hat \beta||_1
Find:
The Ridge Part
The Least Absolute Shrinkage and Selection Operator (LASSO) Part
Why (re)implement glmnet?
Afterall....
- it is standard
- it is fast
However....
- it does not support arbitrary family-link combinations
- it's not designed for out-of-core
- it is written mostly in mortran
mike@Michaels-MacBook-Pro-2:~/projects/glmnet/inst/mortran
> wc -l glmnet5.m
3998 glmnet5.m
mike@Michaels-MacBook-Pro-2:~/projects/glmnet/inst/mortran
> head -n 1000 glmnet5.m | tail -n 20
if g(k).gt.ab*vp(k) < ix(k)=1; ixx=1;>
>
if(ixx.eq.1) go to :again:;
exit;
>
if nlp.gt.maxit < jerr=-m; return;>
:b: iz=1;
loop < nlp=nlp+1; dlx=0.0;
<l=1,nin; k=ia(l); gk=dot_product(y,x(:,k));
ak=a(k); u=gk+ak*xv(k); v=abs(u)-vp(k)*ab; a(k)=0.0;
if(v.gt.0.0)
a(k)=max(cl(1,k),min(cl(2,k),sign(v,u)/(xv(k)+vp(k)*dem)));
if(a(k).eq.ak) next;
del=a(k)-ak; rsq=rsq+del*(2.0*gk-del*xv(k));
y=y-del*x(:,k); dlx=max(xv(k)*del**2,dlx);
>
if(dlx.lt.thr) exit; if nlp.gt.maxit < jerr=-m; return;>
>
jz=0;
>
lewis_glm = function(X, y, lambda, alpha=1, family=binomial, maxit=500,
tol=1e-8, beta=spMatrix(nrow=ncol(X), ncol=1)) {
converged = FALSE
for(i in 1:maxit) {
eta = as.vector(X %*% beta)
g = family()$linkinv(eta)
gprime = family()$mu.eta(eta)
z = eta + (y - g) / gprime
W = as.vector(gprime^2 / family()$variance(g))
beta_old = beta
beta = solve(crossprod(X, W*X), crossprod(X, W*z))
if(sqrt(as.vector(Matrix::crossprod(beta-beta_old))) < tol) {
converged = TRUE
break
}
}
list(beta=beta, iterations=i, converged=converged)
}Start with Bryan Lewis' GLM implementation
pirls = function(X, y, lambda, alpha=1, family=binomial, maxit=500,
tol=1e-8, beta=spMatrix(nrow=ncol(X), ncol=1),
beta_update=coordinate_descent) {
converged = FALSE
for(i in 1:maxit) {
eta = as.vector(X %*% beta)
g = family()$linkinv(eta)
gprime = family()$mu.eta(eta)
z = eta + (y - g) / gprime
W = as.vector(gprime^2 / family()$variance(g))
beta_old = beta
beta = beta_update(X, W, z, lambda, alpha, beta, maxit)
if(sqrt(as.vector(Matrix::crossprod(beta-beta_old))) < tol) {
converged = TRUE
break
}
}
list(beta=beta, iterations=i, converged=converged)
}Make the slope update a function
coordinate_descent = function(X, W, z, lambda, alpha, beta, maxit) {
quad_loss = quadratic_loss(X, W, z, lambda, alpha, beta)
for(i in 1:maxit) {
beta_old = beta
quad_loss_old = quad_loss
beta = update_coordinates(X, W, z, lambda, alpha, beta)
quad_loss = quadratic_loss(X, W, z, lambda, alpha, beta)
if(quad_loss >= quad_loss_old) {
beta = beta_old
break
}
}
if (i == maxit && quad_loss <= quad_loss_old) {
warning("Coordinate descent did not converge.")
}
beta
}
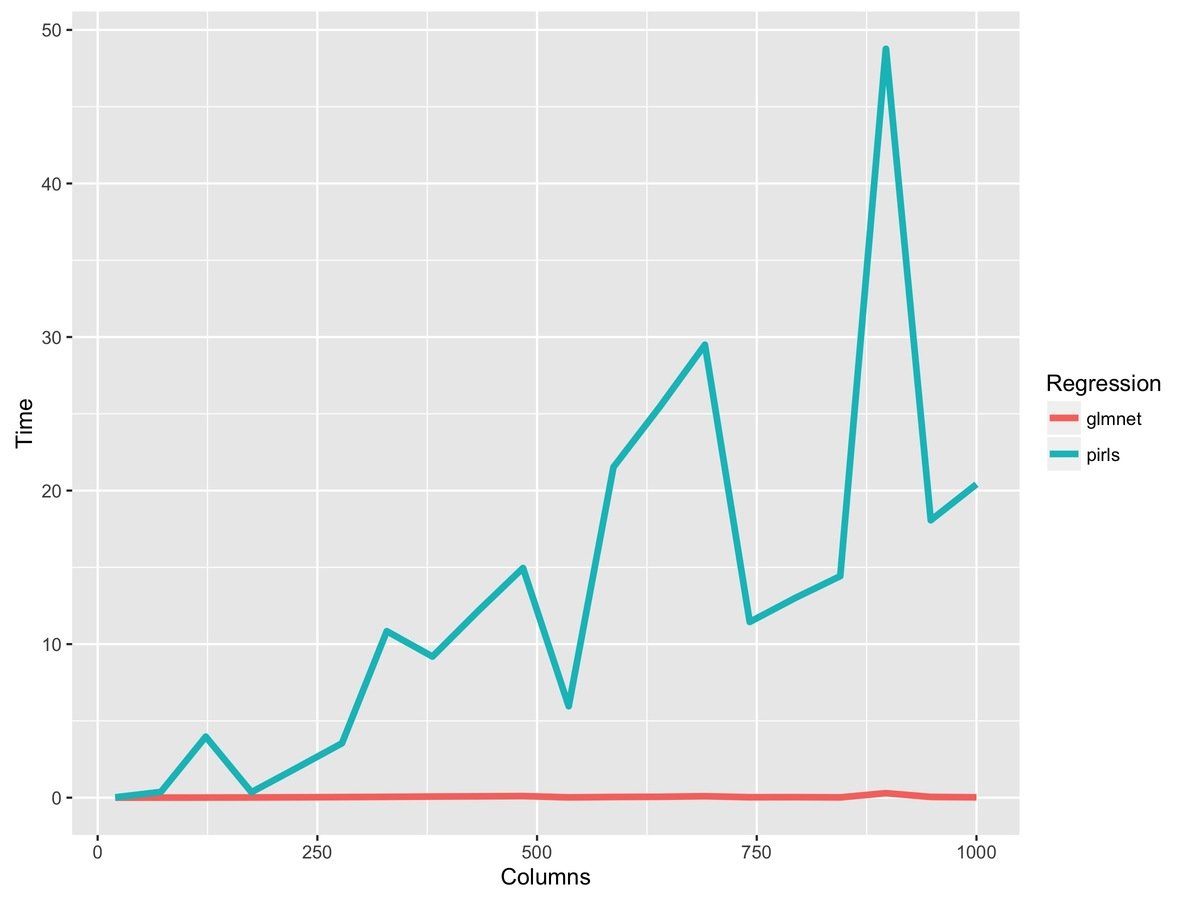
> summaryRprof("pirls_profile.out")
...
$by.total
total.time total.pct self.time self.pct
"beta_update" 1.06 100.00 0.00 0.00
"pirls" 1.06 100.00 0.00 0.00
"update_coordinates" 1.06 100.00 0.00 0.00
Where are spending all of our time?
"Because you're a C++ programmer, there's an above-average chance you're a performance freak. If you're not you're still probably sympathetic to their point of view. (If you're not at all interested in performance, shouldn't you be in the Python room down the hall?)"
-- Scott Meyers, Effective Modern C++
update_coordinates = function(X, W, z, lambda, alpha, beta) {
beta_old = beta
for (i in 1:length(beta)) {
beta[i] = soft_thresh_r(sum(W*X[,i]*(z - X[,-i] %*% beta_old[-i])),
sum(W)*lambda*alpha)
}
beta / (colSums(W*X^2) + lambda*(1-alpha))
}
template<typename ModelMatrixType, typename WeightMatrixType,
typename ResponseType, typename BetaMatrixType>
double update_coordinate(const unsigned int i,
const ModelMatrixType &X, const WeightMatrixType &W, const ResponseType &z,
const double &lambda, const double &alpha, const BetaMatrixType &beta,
const double thresh) {
ModelMatrixType X_shed(X), X_col(X.col(i));
BetaMatrixType beta_shed(beta);
X_shed.shed_col(i);
beta_shed.shed_row(i);
double val = arma::mat((W*X_col).t() * (z - X_shed * beta_shed))[0];
double ret = soft_thresh(val, thresh);
if (ret != 0)
ret /= accu(W * square(X_col)) + lambda*(1-alpha);
return ret;
}
Substitute this...
for this...
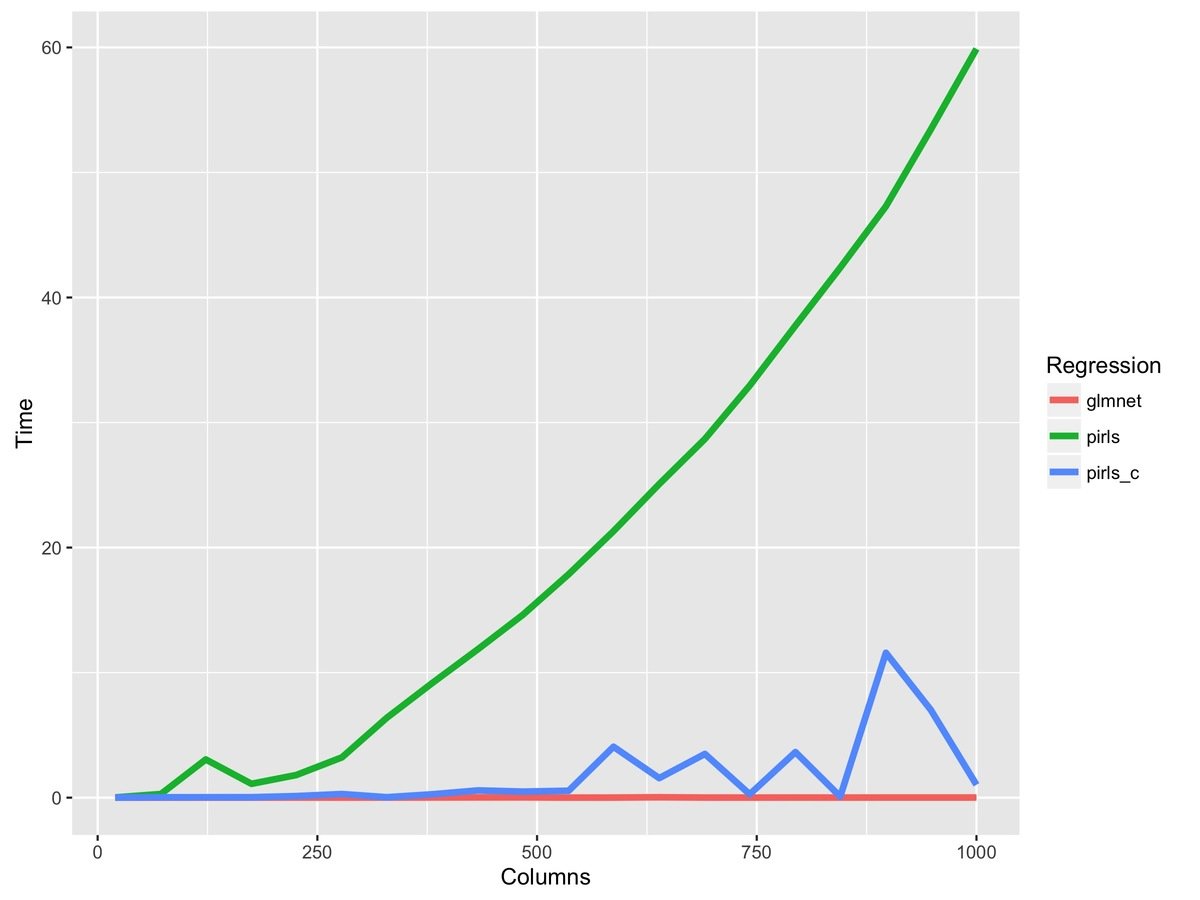
fit_r = pirls(x, y, lambda=lambda, family=gaussian)R code changes from this...
to this...
fit_rc = pirls(x, y, lambda=lambda, family=gaussian,
beta_update=c_coordinate_descent)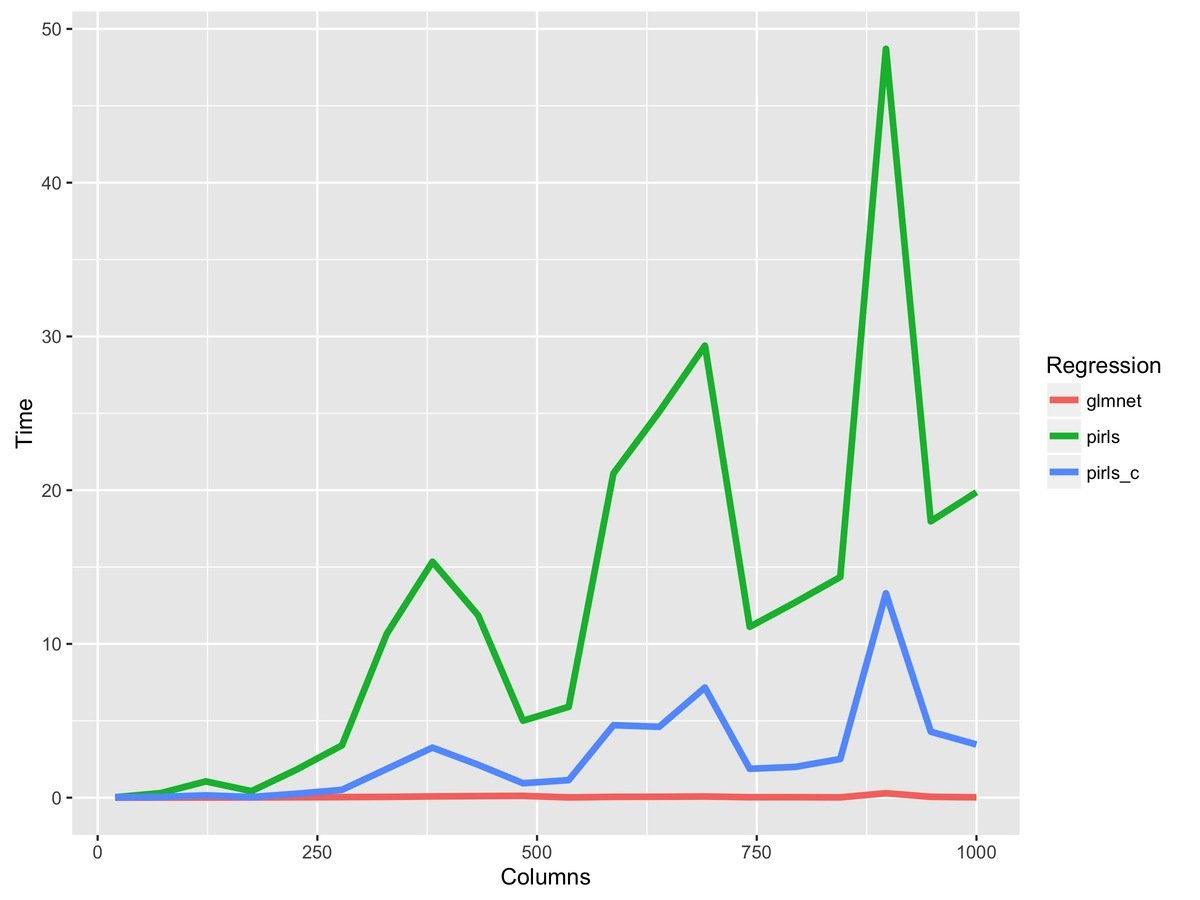
The first rule of fitting high-dimensional models is you don't fit high-dimensional models
Prefilter Regressors
SAFE - discard the jth variable if
|x_j^T y|/n > \lambda - \frac{||x_2|| ||y_2||}{n} \frac{\lambda_{\text{max}} - \lambda}{\lambda_{\text{max}}}
STRONG - discard if KKT conditions are met and
|x_j^T y|/n > 2 \lambda - \lambda_{\text{max}}