Big Data in R
Background for the Course
We are going to explore computational principles for understanding data that may be too big to fit in your computers RAM.
The principles extend to data sets that don't fit on a single machine
These slides are available from slides.com here.
The code is available from github here.
Outline for the Course
Processing Data - transform raw data into something we can analyze
Visualizing Data - gain insight with pictorial representations of data
Fitting Data - say statistically rigorous things about data
Which Data?
This course will make use of the Airline OnTime data set.
- 2009 ASA Stat Computing/Graphics Data Expo
- All U.S. domestic commercial flights for carriers with
at least 1% domestic flights for the year.
- October 1987 - April 2008
- 29 variables per flight (description for each is here)
- ~ 12 gigabytes in size (uncompressed)
Is it "Big"?
It is big enough that it's tough to analyze the whole thing on a single laptop.
It is big enough to show how presented tools scale.
It is not so big that we have to wait a long time for processing/visualizing/fitting.
It is publicly available and small enough for you to download.
Setup: Required Packages
install.packages(c("iotools", "rbokeh",
"tidyr", "foreach",
"datadr", "trelliscope"))
Put the following in R to install the package we'll need.
Part 1: Processing Data
Setup: Get Airline OnTime
url_prefix="http://stat-computing.org/dataexpo/2009/" file_nums = as.character(1987:2008) for (file_num in file_nums) { url = paste0(url_prefix, file_num, ".csv.bz2") new_file_name = paste0(file_num, ".csv.bz2") download.file(url, destfile=new_file_name) }
Exactly how many flights?
file_names = paste0(1987:2008, ".csv.bz2")
system.time({
total_lines = 0
for (file_name in file_names) {
total_lines = total_lines +
nrow(read.csv(bzfile(file_name)))
}
print(total_lines)})
# [1] 123534969
# user system elapsed
# 2619.732 49.698 2671.296
Why is this slow?
1. We are only processing one file at a time.
2. We have to decompress each file.
3. We have to read each file into a data frame.
Process Files in Parallel
library(foreach)
library(doParallel)
cl = makeCluster(4)
registerDoParallel(cl)
system.time({
total_lines = foreach (file_name=file_names,
.combine=`+`) %dopar% {
nrow(read.csv(bzfile(file_name)))
}
print(total_lines)
stopCluster(cl)
})
# [1] 123534969
# user system elapsed
# 0.039 0.107 996.029
Decompress the Files
cl = makeCluster(4) registerDoParallel(cl) foreach (file_name=file_names) %dopar% { x = read.csv(bzfile(file_name)) x[,11] = gsub("'", "", x[,11]) write.csv(x, gsub(".bz2", "", file_name), row.names=FALSE, quote=FALSE) } stopCluster(cl) file_names = gsub(".bz2", "", file_names)
Running time for line count code is now 566.081 seconds.
Use iotools to Read Data
cl = makeCluster(4)
registerDoParallel(cl)
system.time({
total_lines = foreach(file_name=file_names,
.combine=`+`) %dopar% {
library(iotools)
nrow(read.csv.raw(file_name)) } stopCluster(cl) })
Running time for line count code is now 118.206 seconds.
Can we go any faster?
iotools minimizes overhead from "marshalling"
We can use the save and load functions so that there is no marshalling
We can get the row computation in 63.622 seconds.
It doesn't handle when objects are larger than RAM.
The data representation is specific to R.
Why should we care about counting lines of a file?
Our line counter is scalable
It works on manageable subsets of data
It will run faster as more computing resources available
We can extend it to do more than count lines
Is the average flight delay increasing?
It works on manageable subsets of data
It will run faster as more computing resources available
We can extend it to do more than count lines
What if a file is too big to process?
A quick note on pipes
library(tidyr)
add_one = function(x) x+1
add = function(x, y) x+y
3 %>% add_one %>% add(10)
# [1] 14
normalize_df = function(x) { rownames(x) = NULL x$DepTime = sprintf("%04d", x$DepTime) x$DepTime = as.numeric(substr(x$DepTime, 1, 2))*60 + as.numeric(substr(x$DepTime, 3, 4)) x }
file_names = file_names out_file = file("airline.csv", "wb") write(paste(col_names, collapse=","), out_file) for (file_name in file_names) { file_name %>% read.csv.raw %>% normalize_df %>% as.output(sep=",") %>% writeBin(out_file) } close(out_file)
xs = read.csv("1995.csv", nrows=50, as.is=TRUE) col_names = names(xs) col_types = as.vector(sapply(xs, class)) chunk.apply(input="airline.csv", FUN=function(chunk) { nrow(dstrsplit(chunk, col_types, ",")) }, CH.MERGE=sum, parallel=6) # [1] 86289324
airline_apply = function(fun, ch_merge=list,
...,
parallel=6) {
chunk.apply(input="airline.csv",
FUN=function(chunk){
x = dstrsplit(chunk, col_types, ",")
names(x) = col_names
fun(x, ...)
}, CH.MERGE=ch_merge,
parallel=parallel)
}
Let's create some summaries.
Part 2: Visualization
Why do we visualize data?
“There’s nothing better than a picture for making you think of all the questions you forgot to ask” -John Tukey
How do use visualize "big" data
Make the plots interactive.
Make a lot of plots.
An Interactive Visualization Provides
A multi-resolution view of the data
Navigation to the points we are interested in
Extra information for each point
rbokeh
rbokeh
x = read.csv.raw("1995.csv") x = na.omit(x[x$Month==1 & x$Dest=="JFK",]) figure() %>% ly_hist(ArrDelay, data=x, breaks=40, freq=FALSE) %>% ly_density(ArrDelay, data=x)
Arrival vs Departure
figure() %>% ly_points(ArrDelay, DepDelay, data=x, color=UniqueCarrier, hover=list(ArrDelay, DepDelay, UniqueCarrier, Origin))
Single Interactive Plot
Doesn't give us all of the information we want.
Each plot gives us ideas for new plots - so let's make a bunch of plots.
How do we get to the plot we want?
“It seems natural to call such computer guided diagnostics cognostics. We must learn to choose them, calculate them, and use them. Else we drown in a sea of many displays” -John Tukey
Trelliscope
"Trelliscope provides a scalable way to break a set of data into pieces, apply a plot method to each piece, and then arrange those plots in a grid and interactively sort, filter, and query panels of the display based on metrics of interest." - Trelliscope project page
library(trelliscope) x = read.csv.raw("2008.csv") conn = vdbConn("airline-db", name = "airline-db", autoYes=TRUE) bdma = divide(x, c("Month", "DayofMonth", "Dest"))
class(bdma) # [1] "ddf" "ddo" "kvMemory" length(bdma) # [1] 103753 bdma[[1]]$key # [1] "Month= 1|DayofMonth= 1|Dest=ABE" figure() %>% ly_points(ArrDelay, DepDelay, data=bdma[[1]]$value, color=UniqueCarrier, hover=list(ArrDelay, DepDelay, UniqueCarrier, Origin))
Which cognostics should we define?
bdma_cog_fun = function(x) {
list(num_flights=cog(nrow(x), desc="Flights"), num_airlines=cog(length(unique(x$UniqueCarriers)), desc="Carriers"), num_origins=cog(length(unique(x$Origin)), desc="Origins"), avg_dist=cog(mean(x$Distance, na.rm=TRUE), desc="Mean Flight Distance"), sd_dist=cog(sd(x$Distance, na.rm=TRUE), desc="SD Flight Distance"), avg_dep_delay = cog(mean(x$DepDelay, na.rm=TRUE), desc="Mean Dep Delay"), avg_arrival_delay = cog(mean(x$ArrDelay, na.rm=TRUE), desc="Mean Arrival Delay")) }
delay_plot = function(x) { figure() %>% ly_points(ArrDelay, DepDelay, data=x, color=UniqueCarrier, hover=list(ArrDelay, DepDelay, UniqueCarrier, Origin)) } makeDisplay(bdma, name="Delays", group="DayMonthAirport", desc="Delays by Day, Month, and Year", panelFn=delay_plot, cogFn=bdma_cog_fun) view()
Part 3: Fitting (Linear) Models
WHY SHOULD WE CARE ABOUT LINEAR MODELS?
They are old (Gauss 1795, Legendre 1805).
Random Forests (RF) are at least as simple and often do better at prediction.
Deep Learning (DL) dominates vision, speech, voice recognition, etc.
LM generally has lower computational complexity
LM is optimal RF is not
LM can outperform RF for certain learning tasks
RF is more sensitive to correlated noise
LM vs. RF
See most arguments for LM vs RF
We understand characteristics of the space we are in with LM. Not so with DL.
RESULT: We know how things go wrong with LM, not with DL.
LM vs. DL
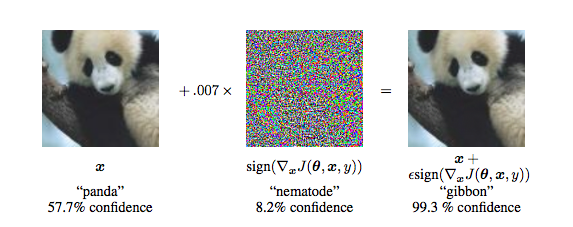
Figure from Explaining and Harnessing Adversarial Examples by Goodfellow et al.
Breaking DL with Adversarial Noise
Advantages of (G)LMs
Computationally Efficient
LM is not simply a prediction tool - results are interpretable
Diagnostics for assessing fit and model assumptions
LM is compatible with other models
> train_inds = sample(1:150, 130)
> iris_train = iris[train_inds,]
> iris_test = iris[setdiff(1:150, train_inds),]
> lm_fit = lm(Sepal.Length ~ Sepal.Width+Petal.Length,
+ data=iris_train)
> sd(predict(lm_fit, iris_test) - iris_test$Sepal.Length)
[1] 0.3879703
> library(randomForest) > iris_train$residual = lm_fit$residuals > iris_train$fitted_values = lm_fit$fitted.values > rf_fit = randomForest( + residual ~ Sepal.Width+Petal.Length+fitted_values, + data=iris_train) > > lm_pred = predict(lm_fit, iris_test) > iris_test$fitted_values = lm_pred > sd(predict(rf_fit, iris_test)+lm_pred - + iris_test$Sepal.Length) [1] 0.3458326
> rf_fit2 = randomForest( + Sepal.Length ~ Sepal.Width+Petal.Length, + data=iris_train) > sd(predict(rf_fit2, iris_test) - iris_test$Sepal.Length) [1] 0.40927
THE NOT SO ORDINARY LEAST SQUARES MODEL
Assume
Find
Derivation
What if X isn't full rank?
my_lm = function(X, y) {
XTy = crossprod(X, y)
XTX = crossprod(X)
QR_XTX = qr(XTX)
keep_vars = QR_XTX$pivot[1:QR_XTX$rank]
solve(XTX[keep_vars, keep_vars]) %*%
XTy[keep_vars]
}
What if X is "almost" not full rank?
my_lm = function(X, y) {
XTy = crossprod(X, y)
XTX = crossprod(X)
QR_XTX = qr(XTX)
keep_vars = QR_XTX$pivot[1:QR_XTX$rank]
solve(XTX[keep_vars, keep_vars]) %*%
XTy[keep_vars]
}
What is X is "almost" not full rank?
Numerical Stability
x1 = 1:100
x2 = x1 * 1.01 + rnorm(100, 0, sd=7e-6)
X = cbind(x1, x2)
y = x1 + x2
qr.solve(t(X) %*% X) %*% t(X) %*% y
# Error in qr.solve(t(X) %*% X) : singular matrix 'a' in solve
beta = qr.solve(t(X) %*% X, tol=0) %*% t(X) %*% y
beta
# [,1]
# x1 0.9869174
# x2 1.0170043
sd(y - X %*% beta)
# [1] 0.1187066
sd(lm(y ~ x1 + x2 -1)$residuals)
# [1] 9.546656e-15
Should I include x1 and x2 if they are colinear?
X = matrix(x1, ncol=1) beta = solve(t(X) %*% X, tol=0) %*% t(X)%*% y beta # [,1] # [1,] 2.01 sd(y - X %*% beta) # [1] 6.896263e-06